The first high-quality chromosome-level genome assembly of the class Cephalopoda was completed by YSFRI
Following the greenfin horse-faced filefish (Thamnaconus septentrionalis), Professor Chen siqing’s reseach group of YSFRI sequenced and assembled the genome of the East Asian common octopus (Octopus sinensis) using PacBio sequencing and Hi-C technology, which is the first chromosome-level genome assembly of the class Cephalopoda around the world. The results were published online recently in Molecular Ecology Resources.
The Cephalopods are a group of highly diverse marine species in the phylum Mollusca distributed worldwide. They have evolved some vertebrate-like biological traits and exhibit complicated behavioral repertoires. Thus, they are interesting species for studying the mechanisms of evolutionary convergence, innovational functional structures, and evolutionary adaptation to a highly active, predatory lifestyle in diverse marine environments. Despite the evolutionary placement and biological significance of cephalopods, genomic data on these organisms remain limited. To date, only five genomes of cephalopod species have been published, including the Octopus vulgaris, Octopus minor, Octopus bimaculoides, Euprymna scolopes and O. sinensis.
The East Asian common octopus, Octopus sinensis (d’Orbigny, 1841) is a valuable commercial species belonging to the class Cephalopoda and lives mainly in shallow temperate waters of the western North Pacific oceans, particularly in the coastal regions of South Korea, China, and Japan. It has long been synonymized with the Atlantic and Mediterranean species, and was previously referred as the ‘common octopus, Octopus vulgaris’. Recently, it was clearly distinguished from O. vulgaris on the basis of molecular and morphological features. O. sinensis is a promising candidate for marine aquaculture because of its high growth rate, large body size (up to 2.5kg) and short life-cycle (about 1 year). In this study, an O. sinensis genome of 2.72 Gb was assembled from 245.01 Gb high-quality PacBio sequences. The assembled genome represents 80.2% completeness (BUSCO) with a contig N50 of 490.36 Kb and a scaffold N50 of 105.89 Mb, showing a considerable improvement compared to other sequenced cephalopod genomes. Hi-C scaffolding of the genome resulted in the construction of 30 pseudo-chromosomes, representing 96.41% of the assembled sequences. The genome contained 42.26% repeat sequences and 5,245 noncoding RNAs. A total of 31,676 protein-coding genes were predicted, of which 82.73% were functionally annotated. Comparative genomic analysis identified 17,020 orthologous gene families, including 819 unique gene families and 629 expanded gene families. This genomic information will be an important molecular resource for further investigation of biological functions and evolutionary adaptations in octopuses, which will also facilitate research into their population genetics and comparative evolution.
YSFRI scientists Li Fenghui, Bian Li and Ge Jianlong of Professor Chen Siqng’s group in YSFRI are the co-first authors, and Professor Chen Siqng is the corresponding author. This work was supported by China Agriculture Research System(CARS-49) and Central Public-interest Scientific Institution Basal Research Fund, CAFS(NO.2020GH02).
Full text link:https://doi.org/10.1111/1755-0998.13216

Figure 1 The East Asian common octopus (Octopus sinensis)

Figure 2 The genome-wide Hi-C map of O. sinensis
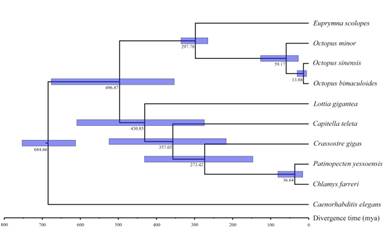
Figure 3 The phylogenetic analysis of the O. sinensis and other nine bilaterian species
